Cloud-Optimized HDF/NetCDF
The following provides guidance on how to assess and create cloud-optimized HDF5 and NetCDF4 files. Cloud-optimized formats provide efficient subsetting without maintaining a service, such as OpenDAP, Hyrax or SlideRule. Often HDF5 and NetCDF-4 formats are a requirement, for archival purposes. If HDF5 and NetCDF-4 formats are not a requirement, a cloud-native format like Zarr should be considered. If HDF5/NetCDF-4 formats are a requirement, consider zarr-readable chunk indexes such as kerchunk and VirtualiZarr.
You can skip the background and details by jumping to the checklist.
Background
NetCDF and HDF were originally designed with disk access in mind. As Matt Rocklin explains in HDF in the Cloud: Challenges and Solutions for Scientific Data:
The HDF format is complex and metadata is strewn throughout the file, so that a complex sequence of reads is required to reach a specific chunk of data.
Why accessing HDF5 on the cloud is slow
In the diagram below, R0, ..., Rn represent metadata requests. A large number of these requests slows down working with these files in the cloud.
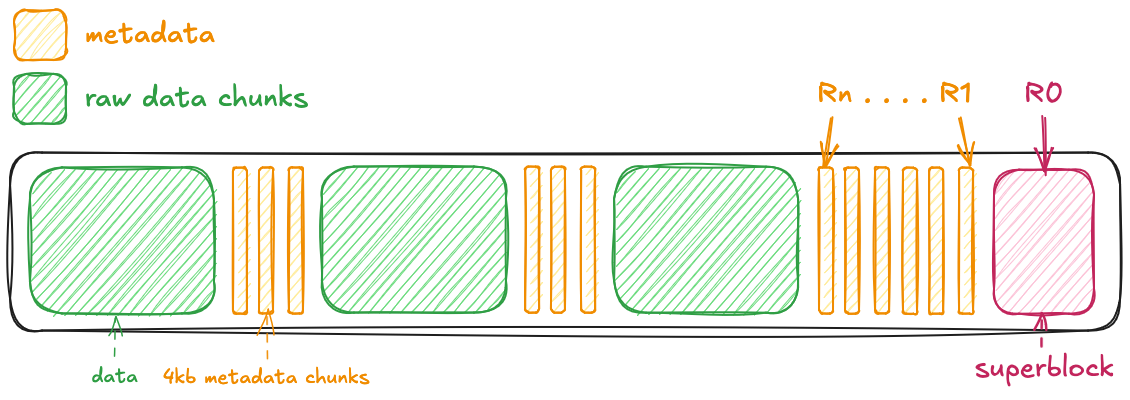
When reading and writing data from disk, small blocks of metadata and raw data chunks were preferred because access was fast, and retrieving any part of a chunk involved reading the entire chunk (H5py Developers n.d.). However, when this same data is stored in the cloud, performance can suffer due to the high number of requests required to access both metadata and raw data. With network access, reducing the number of requests makes access much more efficient.
A detailed explanation of current best practices for cloud-optimized HDF5 and NetCDF-4 is provided below, followed by a checklist and some how-to guidance for assessing file layout.
Note: NetCDF-4 are valid HDF5 files, see Reading and Editing NetCDF-4 Files with HDF5.
Current Best Practices for Cloud-Optimized HDF5 and NetCDF-4
Format
To be considered cloud-optimized, the format should support chunking and compression. NetCDF3 and HDF4 prior to v4.1 do not support chunking and chunk-level compression, and thus cannot be reformatted to be cloud optimized. The lack of support for chunking and compression along with other limitations led to the development of NetCDF-4 and HDF5.
Consolidated Internal File Metadata
Consolidated metadata is a key characteristic of cloud-optimized data and enables “lazy loading” (see the Lazy Loading block below). Client libraries use file metadata to understand what’s in the file and where it is stored. When metadata is scattered across a file (which is the default for HDF5 writing), client applications have to make multiple requests for metadata information.
For HDF5 files, to consolidate metadata, files should be written with the paged aggregation file space management strategy (see also H5F_FSPACE_STRATEGY_PAGE). When using this strategy, HDF5 will write data in pages where metadata is separated from raw data chunks. Note the page size should also be set, as the default size is 4096 bytes (or 4KB, source). Further, only files using paged aggregation can use the HDF5 page buffer cache – a low-level library cache (Jelenak 2022) – to reduce subsequent data access.
Lazy loading is a common term for first loading only metadata, and deferring reading of data values until required by computation.
HDF5 file organization—data, metadata, and free space—depends on the file space management strategy. Details on these strategies are in HDF Support: File Space Management.
Here are a few additional considerations for understanding and implementing the H5F_FSPACE_STRATEGY_PAGE strategy:
- Chunks vs. Pages: In HDF5, datasets can be chunked, meaning the dataset is divided into smaller blocks of data that can be individually compressed (see also Chunking in HDF5). Pages, on the other hand, represent the smallest unit HDF5 uses for reading and writing data. To optimize performance, chunk sizes should ideally align with the page size or be a multiple thereof. Entire pages are read into memory when accessing chunks or metadata. Only the relevant data (e.g., a specific chunk) is decompressed.
- Page Size Considerations: The page size applies to both metadata and raw data. Therefore, the chosen page size should strike a balance: it must consolidate metadata efficiently while minimizing unused space in raw data chunks. Excess unused space can significantly increase file size. File size is typically not a concern for I/O performance when accessing parts of a file. However, increased file size can become a concern for storage costs.
Chunk Size
As described in Chunking in HDF5, datasets in HDF5 can be split into chunks and stored in discrete compressed blocks.
How to determine chunk size
The uncompressed chunk size is calculated by multiplying the chunk dimensions by the size of the data type. For example, a 3-dimensional chunk with dimension lengths 10x100x100 and a float64 data type (8 bytes) results in an uncompressed chunk size of 0.8 MB.
When designing chunk size, usually the size is for the uncompressed chunk. This is because:
- Data variability: Because of data variability, you cannot deterministically know the size of each compressed chunk.
- Memory considerations: The uncompressed size determines how much memory must be available for reading and writing each chunk.
How to choose a chunk size
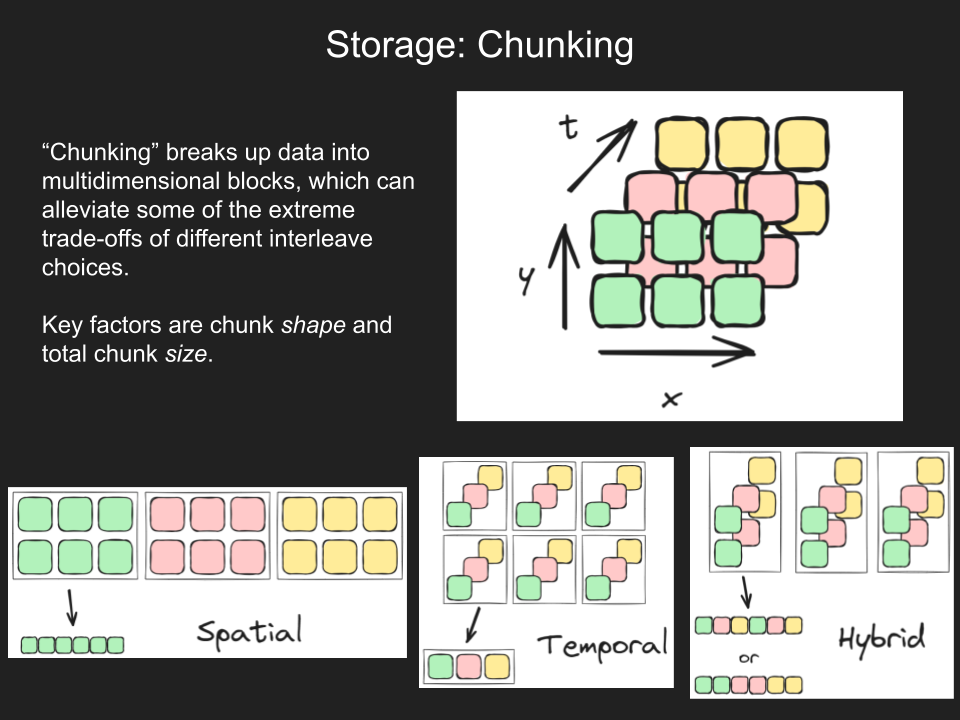
There is no one-size-fits all chunk size and shape as files, use cases, and storage systems vary. However, chunks should not be “too big” or “too small”.
When chunks are too small:
- Extra metadata may increase file size.
- It takes extra time to look up each chunk.
- More network I/O is incurred because each chunk is stored and accessed independently (although contiguous chunks may be accessed by extending the byte range into one request).
When chunks are too big:
- An entire chunk must be read and decompressed to read even a small portion of the data.
- Managing large chunks in memory slows down processing and is more likely to exceed memory and chunk caches.
A chunk size should be selected that is large enough to reduce the number of tasks that parallel schedulers have to think about (which affects overhead) but also small enough so many can fit in memory at once. The Amazon S3 Best Practices says the typical size for byte-range requests is 8-16MB. However, requests for data from contiguous chunks can be merged into 1 HTTP request, so chunks could be much smaller (one recommendation is 100kb to 2mb) (Jelenak 2024).
Performance greatly depends on libraries used to access the data and how they are configured to cache data as well.
Chunk shape vs chunk size
The chunk size must be differentiated from the chunk shape, which is the number of values stored along each dimension in a given chunk. Recommended chunk size depends on a storage system’s (such as S3) characteristics and its interaction with the data access library.
In contrast, an optimal chunk shape is use case dependent. For a 3-dimensional dataset (latitude, longitude, time) with a chunk size of 1000, chunk shapes could vary, such as:
- 10 lat x 10 lon x 10 time,
- 20 lat x 50 lon x 1 time, or,
- 5 lat x 5 lon x 40 time.
Larger chunks in a given dimension improve read performance in that direction: (3) is best for time-series analysis, (2) for mapping, and (1) is balanced for both. Thus, chunk shape should be chosen based on how the data is expected to be used, as there are trade-offs. A useful approach is to think in terms of the chunks’ aspect ratio, adjusting relative dimension lengths to fit the desired optimization for spatial versus time-series analyses (see https://github.com/jbusecke/dynamic_chunks).
A best practice to help determine both chunk size and shape would be to specify some “benchmark use cases” for the data. With these use cases in mind, evaluate what chunk shape and size is large enough such that the computation doesn’t result in thousands of jobs and small enough that multiple chunks can be stored in-memory and a library’s buffer cache, such as HDF5’s buffer cache.
Additional chunk shape and size resources
- Unidata Blog: “Chunking Data: Choosing Shapes”
- HDF Support Site: “Chunking in HDF5”
- The dynamic_chunks module by Julius Busecke may help in determing a chunk shape based on a target size and dimension aspect ratio.
Compression
Compression is the process of minimizing the size of data stored using an algorithm which can condense data through various methods. This can include scale and offset parameters which reduce the size of each byte that needs to be stored. There are many algorithms for compressing data and users can even define their own compression algorithms. Data product owners should evaluate what compression algorithm is right for their data.
NASA satellite data is predominantly compressed with the zlib (a.k.a., gzip, deflate) method. However, other methods should be explored as a higher compression ratio is often possible, and in the case of HDF5, fills file pages better(Jelenak 2023a).
Data Usage Documentation through Tutorials and Examples
Tutorials and examples are starting points for many data users. These documents should include information on how to read data directly from cloud storage (as opposed to downloading over HTTPS) and how to configure popular libraries for optimizing performance.
For example, the following library defaults will impact performance and may be included in data usage documentation:
- HDF5 library:
- Chunk cache: The size of the HDF5’s chunk cache by default is 1MB. This value is configurable. Chunks that don’t fit into the chunk cache are discarded and must be re-read from the storage location each time. See also: Improve HDF5 performance using caching and h5py documentation: Chunk cache.
- Page buffer size: The H5Pset_page_buffer_size and the page_buf_size argument to h5py.File should match up with the page size to optimize reading the data in pages.
- S3FS library: The S3FS library is a popular library for accessing data on AWS’s cloud object storage S3. Note that S3FS has a default block size, which may change, so please refer to the S3FS API docs for the default setting. And note it can be set upon instantiation of an S3FileSystem object.
- Additional guidance on h5py, fsspec, and ROS3 libraries for creating and reading HDF5 can be found in Jelenak (2024).
Additional research
Here is some additional research done on caching for specific libraries and datasets that may be helpful in understanding the impact of caching and developing product guidance:
- In this issue Optimize s3fs read cache settings for the GEDI Subsetter (findings to be formalized), Chuck Daniels found the “all” cache type (cache entire contents), a block size of 8MB and fill cache=True to deliver the best performance. NOTE: This is for non-cloud-optimized data.
- In HDF at the Speed of Zarr, Luis Lopez demonstrates, using ICESat-2 data, the importance of using similar arguments with fsspec (blockcache instead of all, but the results in the issue above were not significantly different between these 2 options) as well as the importance of using nearly equivalent arguments in for h5py (raw data chunk cache nbytes and page_buff_size).
Cloud-Optimized HDF/NetCDF Checklist
Please consider the following when preparing HDF/NetCDF data for use on the cloud:
How tos
The examples below require the HDF5 library package is installed on your system. These commands will also work for NetCDF-4. While you can check for chunk size and shape with h5py, h5py is a high-level interface primarily for accessing datasets, attributes, and other basic HDF5 functionalities. h5py does not expose lower-level file options directly.
Commands in brief:
How to check for consolidated metadata
To be considered cloud-optimized, HDF5 files should be written with the PAGE file space management strategy (see also File Space Management Strategies). When using this strategy, HDF5 will write aggregate metadata and raw data into fixed-size pages (Jelenak 2023b).
You can check the file space management strategy with the command line h5stat tool:
h5stat -S infile.h5This returns output such as:
Filename: infile.h5
File space management strategy: H5F_FSPACE_STRATEGY_FSM_AGGR
File space page size: 4096 bytes
Summary of file space information:
File metadata: 2157376 bytes
Raw data: 37784376 bytes
Amount/Percent of tracked free space: 0 bytes/0.0%
Unaccounted space: 802424 bytes
Total space: 40744176 bytesNotice the strategy: File space management strategy: H5F_FSPACE_STRATEGY_FSM_AGGR. This is the default option. The best choice for cloud-optimized access is H5F_FSPACE_STRATEGY_PAGE. Learn more about the options in the HDF docs: File Space Management (HDF Group).
How to change the file space management strategy
You can use the h5repack to reorganize the metadata (Jelenak 2023b). When repacking to use the PAGE file space management strategy, you will also need to specify a page size that will indicate the block size for metadata storage. This should be at least as big as the File metadata value returned from h5stat -S.
$ h5repack -S PAGE -G 4000000 infile.h5 outfile.h5When reading the HDF5 library needs to be configured to use the page aggregated files. If using the HDF5 library you can set H5Pset_page_buffer_size. For h5py File objects you can set page_buf_size when instantiating the File object.
- h5repack’s aggregation is fast but rechunking is slow. You may want to use the h5py library directly to repack. See an example of how to do so in NSIDC’s cloud-optimized ICESat-2 repo: optimize-atl03.py.
- The NetCDF library doesn’t expose the low-level HDF5 API so one must first create the file with the NetCDF library and then repack it with h5repack or python. You can also create NetCDF files with HDF5 libraries, however required NetCDF properties must be set. See also: Using the HDF5 file space strategy property Unidata/netcdf-c #2871.
author credit: Luis Lopez
How to check chunk size and shape
Replace infile.h5 with a filename on your system and dataset_name with the name of a dataset in that file.
h5dump -pH infile.h5 | grep dataset_name -A 10-p shows the properties (dataset layout, chunk size and compression) of objects in the HDF5 file. -H prints the header information only. Together, we get metadata about the structure of the file.
Example command with output and how to read it, in comments:
$ h5dump -pH ~/Downloads/ATL03_20240510223215_08142301_006_01.h5 | grep h_ph -A 10
DATASET "h_ph" {
DATATYPE H5T_IEEE_F32LE # Four-byte, little-endian, IEEE floating point
DATASPACE SIMPLE { ( 15853996 ) / ( H5S_UNLIMITED ) } # simple (n-dimensional) dataspace, this is a 1-dimensional array of length 15,853,996
STORAGE_LAYOUT {
CHUNKED ( 10000 ) # Dataset is stored in chunks of 10,000 elements per chunk
SIZE 57067702 (1.111:1 COMPRESSION) # Compressed datasets compression ratio of 1.111 to 1 - only slightly compresed
}
FILTERS {
COMPRESSION DEFLATE { LEVEL 6 } # Compressed with deflate algorithm with a compression level of 6
}
...How to change the chunk size and shape
$ h5repack \
# dataset:CHUNK=DIM[xDIM...xDIM]
/path/to/dataset:CHUNK=2000 infile.h5 outfile.h5Closing Thoughts
Many existing HDF5 and NetCDF4 collections use the default file space management strategy and with very small raw data chunks. While optimizing these collections would improve performance, it requires significant effort, including benchmark design and development, reprocessing and a deep understanding of HDF5 file space management and caching. When time and resources are limited, tools like Kerchunk and VirtualiZarr offer a practical alternative. These tools don’t rechunk the data but instead consolidate metadata, resulting in notable performance improvements.